Introduction
Polycystic ovary syndrome (PCOS) is a metabolic and reproductive disorder affecting 5%-10% of women of reproductive age (1). It is characterized by increased ovarian and adrenal androgen secretion, hyperandrogenic symptoms (hirsutism, acne, alopecia), menstrual irregularities and insulin resistance (2), which is present in 50%-70% of PCOS patients. In addition to insulin resistance, other features of metabolic syndrome are often also present (obesity, dyslipidemia, hypertension and impaired glucose tolerance). Besides PCOS and metabolic syndrome, insulin resistance is also associated with obesity, type 2 diabetes mellitus, lipodystrophies and chronic infection. The overall prevalence of insulin resistance is reported to be 10%-25% (1). Insulin resistance is characterized by attenuated response of tissues to insulin action and can be due to impairments at any level of insulin signaling. Clinical markers of insulin resistance are visceral obesity, acanthosis nigricans, acne, hirsutism and hepatic steatosis. Hyperinsulinemic euglycemic clamp is considered to be the gold standard for evaluating insulin resistance. Because of the invasiveness of this method, other alternative measures like fasting plasma insulin concentration, homeostasis model assessment (HOMA) index, quantitative insulin sensitivity check index (QUICKI) and McAuley index have also been developed (1).
Peroxisome proliferator-activated receptors (PPARs) are nuclear receptors which control gene transcription. There are three types of PPARs: PPARα, PPARβ (also named PPARδ) and PPARγ. PPARγ has an important role as an adipocyte gene expression regulator. Specific PPARγ activation induces preadipocyte differentation into mature adipocyte in rodent and human cell lines (3). PPARγ regulates transcription of proteins involved in free fatty acid (FFA) uptake (adipocyte-specific FFA binding protein aP2, lipoprotein lipase, fatty acids transport proteins FATP and CD36, adipophilin, liver X receptor α) and lipogenesis (long chain acyl-CoA synthase, phosphoenolpyruvate carboxykinase) (4). Besides adipocyte differentiation, PPARγ is also essential for mature adipocyte survival in vitro (5).
PPARG gene is located on chrosomome 3p25. It spans over 146 kilobase pairs and consists of 9 exons. It codes 4 different mRNAs which translate to 2 different protein isoforms (PPARγ1 and PPARγ2) using different promotors and alternative splicing. The PPARγ2 isoform has 30 additional amino acids on its N-terminal end and has 5- to 6-fold increase in its ligand-dependent activating function (5). The Pro12Ala polymorphism lies in exon B and is therefore present only in PPARγ2 isoform. The His477His polymorphism lies in exon 6 and can be found in both protein isoforms (Figure 1). PPARγ containing the G allele has a lower transcriptional activity than the one containing the allele C in vitro. This was confirmed by 3 individual in vivo studies (7).
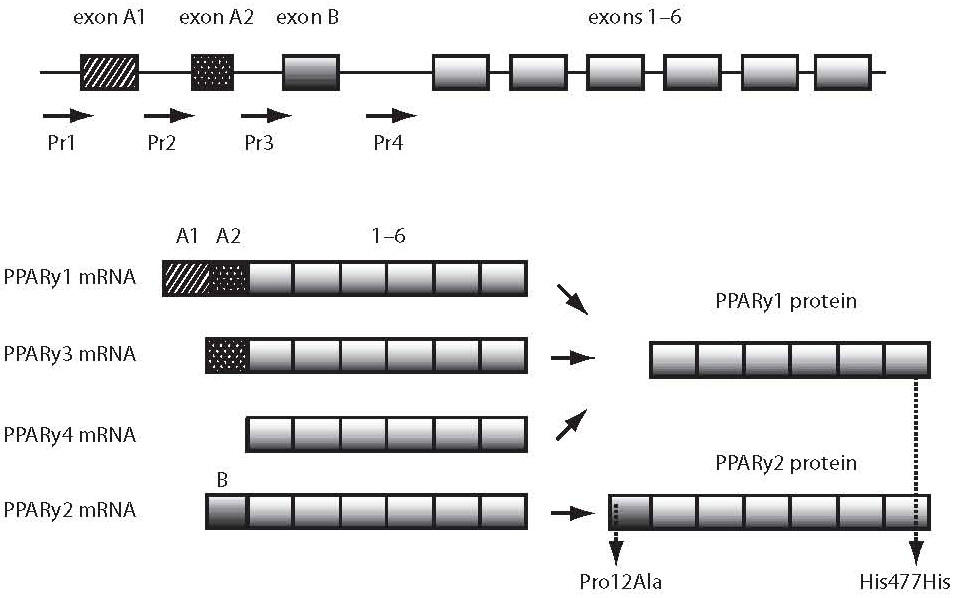
Figure 1. PPARγ gene and mRNAs. The PPARγ1 isoform is 475 amino acids long and is a result of translation of PPARγ1, PPARγ3 and PPARγ4 mRNA. The PPARγ2 isoform has 30 additional amino acids on its N-terminal end. PPARγ1 is expressed ubiquitously, whereas PPARγ2 is adipose-specific. PPARγ3 is expressed in macrophages, adipose tissue and colon. Localization of PPARγ4 expression is not yet defined. (Adapted from ref. 6)
The aim of our preliminary study was to determine the association of Pro12Ala and His477His polymorphisms in PPARG gene and their diplotypes with insulin resistance in PCOS patients. Although many studies have studied the Pro12Ala and/or His477His polymorphisms, often with opposing results, we were the first to conduct this kind of a study in a Slovene group of PCOS patients and to determine the association of diplotype with high sensitivity C-reactive protein (hsCRP) in PCOS patients.
Patients and methods
Patients
Our study included 69 Slovene (Caucasian) women, all of them diagnosed with PCOS according to NICHD (National Institute of Child Health and Human Development) criteria. The study was approved by the Ethics Committee and all patients signed an informed consent.
Methods
Anthropometric measurements included body weight, height and waist circumference. Subject systolic and diastolic blood pressures were measured using routine methods. Peripheral blood was drawn in fastening state for genetic studies and measurement of biochemical parameters. Glucose was also measured after oral glucose tolerance test (OGTT) using GOD-PAP method (Roche Hitachi 917, Roche, Mannheim, Germany). Chemiluminescent immunochemical method was used for insulin (Liaison Insulin, Diasorin, Saluggia VC, Italy) and hsCRP (Immulite, DPC Inc., Los Angeles, USA) measurements. All parameters were measured in a certified laboratory using routine validated methods. HOMA index was calculated from the findings using the following equation:
HOMA-IR = fasting insulin (mU/L) × x fasting glucose (mmol/L)/22.5 (8).
DNA was isolated from peripheral blood using FlexiGene DNA Kit (Qiagen, Hilden, Germany). Genotypes were determined using PCR-RFLP methods. The Pro12Ala polymorphism was genotyped using mismatch primers and restriction enzyme HpaII (New England Biolabs) previously described by Globočnik et al. (9). The His477His polymorphism (also named C161T, C1431T, His447His and CAC477CAT) was genotyped using primers PPARG-Ex6_F: CAGGTTTGCTGAATGTGAAGC and PPARG-Ex6_R: TGGCTCAGGACTCTCTGCTAGT (designed using Primer3 software) and restriction enzyme NlaIII (New England Biolabs). Negative control was always used in RFLP genotyping and duplicate analysis were performed on at least 10% of samples.
Statistical analysis
Hardy-Weinberg equilibrium was determined using Courtlab calculator (10). Data were analyzed with SPSS 13.0 for Windows. A P > 0.05 was considered statistically significant. Data distribution was tested using Kolmogorov-Smirnov test. Genotype association with clinical markers was tested using Student’s t-test for variables with normal distribution or Mann-Whitney test for variables that didn’t show normal distribution. Haplotype frequencies were calculated with LDA 1.0 program (11). Haplotypes were assigned using HAP program (12). Diplotype association with clinical markers with homogenic variance was analyzed using ANOVA and post-hoc tests (Bonferroni, Scheffe, Tukey HSD). Diplotype association with clinical markers with non-homogenic variance was analyzed with Kruskal-Wallis test.
Results
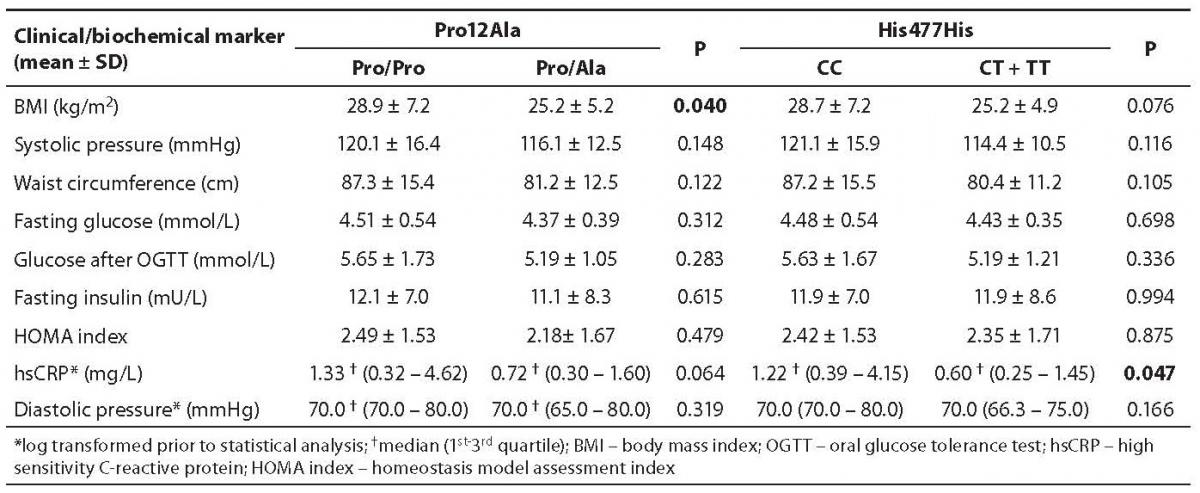
Genotype frequencies for both SNPs were in Hardy-Weinberg equilibrium in our study group. Genotype and allele frequencies are shown in Table 1. Thirty-four PCOS patients had insulin resistance according to HOMA index above 2.0. A significant difference was found between Pro12Ala genotype subgroups according to body mass index (BMI) (Table 2), where carriers of the G allele had lower BMI (Pro/Pro BMI = 28.9 ± 7.2 kg/m2 vs. C/T BMI = 25.2 ± 5.2 kg/m2; P = 0.040). The Pro12Ala polymorphism was not associated with systolic or diastolic blood pressure, waist circumference, fasting glucose, glucose after OGTT, fasting insulin, HOMA index or hsCRP.
Table 1. Allele and genotype frequencies of Pro12Ala and His477His polymorphisms in polycystic ovary syndrome (PCOS) patients
A significant difference was also found for the His477His polymorphism according to hsCRP (Table 2). Carriers of the T allele had lower hsCRP and therefore a lower risk of developing cardiovascular disease (data displayed as median (1st quartile-3rd quartile); C/C hsCRP = 1.22 (0.39-4.15) mg/L vs. C/T + T/T hsCRP = 0.60 (0.25-1.45) mg/L; P = 0.047). The His477His polymorphism was not associated with BMI, systolic or diastolic blood pressure, waist circumference, fasting glucose, glucose after OGTT, fasting insulin or HOMA index.
Table 2. Clinical and biochemical parameters in Pro12Ala and His477His polymorphism subgroups
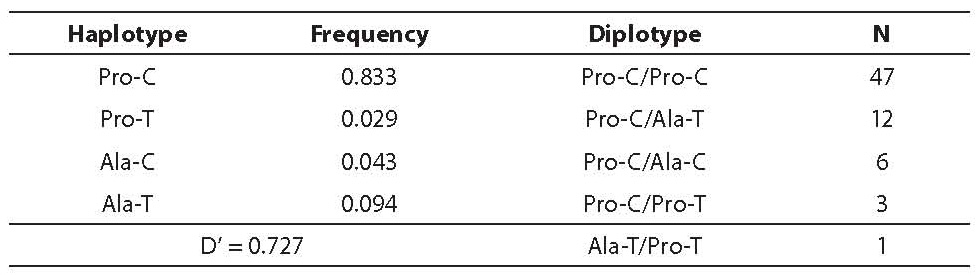
In addition, haplotype analysis was performed. The Pro12Ala and His477His polymorphisms were in considerable linkage disequilibrium (D’ = 0.727) (Table 3). Out of ten theoretically possible diplotypes, only five diplotypes were found. Only one subject had the Ala-T/Pro-T diplotype and was excluded from further statistical analysis. For the rest, diplotype association was tested for clinical and biochemical markers of insulin resistance, yielding no significant association (data not presented).
Table 3. Haplotype frequencies and diplotypes
Discussion
We found significant association between the G allele of the Pro12Ala polymorphism and lower BMI, and between the T allele of the His477His polymorphism and lower hsCRP but none of the two polymorphisms was associated with fasting plasma insulin and glucose concentrations and HOMA index. Diplotype analysis showed no association with anthropometric and biochemical markers of insulin resistance in PCOS.
Pro12Ala is a common polymorphism; G allele frequency is race dependent and spans from 1% to 16%. Our results in PCOS patients showed an allele frequency of 13.8% for the G allele. These frequencies are similar to those established in Danes (13), Scots (14,15), Finns (16), French (17), Indians (18), Americans (19) and Slovenes (9). A lower allele frequency (8%) was found in USA Caucasian females, and between 1% and 4% in African Americans (20), Chinese, Malayans (18), Koreans (21) and Italian females (22). A number of studies explored the influence of the G allele on tissue insulin sensitivity. Some showed the G allele to improve tissue insulin sensitivity, some failed to find any connection and, interestingly, none found a negative impact of the G allele. Results are even more inconsistent regarding diabetes. Some studies linked the G allele with a decreased risk, some with an increased risk of type 2 diabetes and some found no connection (7).
The His477His polymorphism can be found under different names: C161T, C1431T, His447His and CAC477CAT. The T allele frequency ranges from 6.5% to 25.2%, depending on race. The determined T allele frequency for His477His polymorphism in our study was 12.3% in PCOS patients. Similar allele frequencies were found in French (17,23), Americans (19) and Scots (14,15). Slightly higher allele frequencies (16%-17%) were found in Australians (24) and Indians, and even higher (19%-25%) in Chinese, Malayans (18) and Finns (16).
Statistical analysis showed a significant association between the G allele of the Pro12Ala polymorphism and BMI where the carriers of the G allele had lower BMI. Some of the published correlation studies showed no significant association of the G allele with BMI (13,21), three of them including women with PCOS (20,22,25). Others found the G allele carriers to have higher BMI (16,18,19). All of these studies were conducted on a 2- to 60-fold larger number of subjects compared to our study. A meta-analysis including data from 30 studies (~19000 subjects) showed a significant connection of the G allele in subjects with BMI over 27 kg/m2 (26). Another meta-analysis including data from 57 studies (~32000 subjects) showed the Pro12Ala to have no association with diabetes related traits but in certain subgroups (Caucasians, obese subjects) the G allele was associated with higher BMI and greater insulin sensitivity (27).
In our study, the Pro12Ala polymorphism was not associated with waist circumference and diastolic and systolic blood pressure, which is consistent with the findings of Frederiksen et al. (13) but inconsistent with Valve et al. and Wei et al. (16,19), where the G allele was connected with higher waist circumference and higher BMI as well. Like our study, none of those studies (13,16,21,22,25) found any association between the G allele and fasting glucose, fasting insulin and/or HOMA index. Hara et al. (20) found no significant association of the G allele and fasting glucose or glucose after OGTT, but carriers of the G allele had a significantly lower insulin concentration and HOMA index. Similar to our study, the G allele was not associated with fasting insulin in studies conducted by Tai et al. (18) and Wei et al. (19), but it was associated with higher and lower glucose, respectively. Studies of the Pro12Ala polymorphism in association with hsCRP in PCOS patients cannot be found in the literature.
Our statistical analysis on the His477His polymorphism showed a significant association with hsCRP; carriers of the T allele had lower hsCRP. Similar to Meirhaeghe et al. (23), we found no association of the T allele with BMI. In contrast to our findings, the T allele was associated with higher BMI in several studies (16,18,19,22). In our study, the T allele was not associated with waist circumference, which is similar to the results reported by Wei et al. (19). In the study by Valve et al. (16), it was associated with higher waist circumference (16), which is in contrast to our results. Meirhaeghe et al. (23) found no association between the T allele and systolic and diastolic blood pressure, which is consistent with our findings. Fasting glucose and insulin concentrations were not associated with the T allele in our study, which is similar to the findings reported elsewhere (16,18,19,22,23). We showed the T allele to be associated with decreased hsCRP, therefore representing a lower risk for cardiovascular disease development. Similar findings have been reported by Wang et al., where carriers of the T allele had a lower risk of developing cardiovascular disease, an association seen especially in CT heterozygotes (24).
We performed a diplotype analysis and established the degree of linkage disequilibrium D’ = 0.727, which was similar to the one reported by Doney et al. in diabetic Scots (D’ = 0.697) (14). Somewhat lower D’s were found in another study in diabetic Scots (D’ = 0.663), healthy children (D’ = 0.638) and healthy adults (D’ = 0.608) (15), Caucasian Americans (D’ = 0.65) (19), Chinese (D’ = 0.555) and Malayans (D’ = 0.572). A higher degree of linkage disequilibrium was found in Indians (D’ = 0.799) (18). Haplotype frequencies in our subjects (Table 3) were similar to those found by Doney et al. in Scots in two studies (14,15).
Our results showed that diplotype was not associated with any of the anthropometric or biochemical markers. On the contrary, Doney et al. demonstrated that Pro-T haplotype was connected with higher BMI compared to Pro-C or Ala-C haplotype in diabetic and non-diabetic Scots (14).
Considering the fact that the results of several studies are inconsistent, it is possible that the Pro12Ala and His477His polymorphisms only have a small influence on insulin resistance. In our pilot study on individual polymorphisms we showed that carriers of the G allele of the Pro12Ala polymorphism had a lower BMI and carriers of the T allele of the His477His polymorphism had a lower hsCRP, which are two risk factors for the development of insulin resistance. However, the association with the HOMA index or fasting insulin, two of insulin resistance markers, was not observed in our study. We therefore conclude that the polymorphisms investigated are not associated with insulin resistance in PCOS patients.
Acknowledgment
This study was supported by Slovenian Research Agency, Research Program No. P3-0298.
Notes
Potential conflict of interest
None declared
References
1. Mlinar B, Marc J, Janež A, Pfeifer M. Molecular mechanisms of insulin resistance and associated diseases. Clin Chim Acta 2007;375:20-35.
2. Escobar-Morreale HF, Luque-Ramirez M, San Millan JL. The molecular-genetic basis of functional hyperandrogenism and the polycystic ovary syndrome. Endocr Rev 2005;26:251-82.
3. Ferre P. The biology of peroxisome proliferator-activated receptors: relationship with lipid metabolism and insulin sensitivity. Diabetes 2004:53:S43-S50.
4. Cock TA, Houten SM, Auwerx J. Peroxisome proliferator-activated receptor-γ: too much of a good thing causes harm. EMBO Rep 2004;5:142-7.
5. Knouff C, Auwerx J. Peroxisome proliferator-activated receptor- calls for activation in moderation: lessons from genetics and pharmacology. Endocr Rev 2004;25:899-918.
6. Merihaeghe A, Fajas L, Gouilleux F, Cottel D, Helbecque N, Auwerx J, et al. A functional polymorphism in a STAT5B site of the human PPARγ3 gene promoter affects height and lipid metabolism in a French population. Arterioscler Thromb Vasc Biol 2003;23:289-94.
7. Meirhaeghe A, Amouyel P. Impact of genetic variation of PPARγ in humans. Mol Genet Metab 2004;83:93-102.
8. Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC. Homeostasis model assessment: insulin resistance and ß cell function from fasting plasma glucose and insulin concentration in man. Diabetologia 1985;28:412-9.
9. GlobočnikPetrovič M, KunejT, PeterlinB, Dovč P, Petrovič D. Gly482Serpolymorphismoftheperoxisomeproliferator-activatedreceptor-γcoactivator-1 genemightbeariskfactorfordiabeticretinopathyinSlovenepopulation (Caucasians) withtype 2diabetesandthePro12AlapolymorphismofPPARγgeneisnot. Diabetes Met Res Rev 2005;21:470-4.
11. Ding K, Zhou K, He F, Shen Y. LDA - a java-based linkage disequilibrium analyzer. Bioinformatics 2003;19:2147-8.
12. Halperin E, Eskin E. Haplotype reconstruction from genotype data using imperfect phylogeny. Bioinformatics 2004;20:1842-9.
13. Frederiksen L, Brǿdbǽk K, Fenger M, Jǿrgensen T, Borch-Johnsen K, Madsbad SM, et al. Studies of the Pro12Ala polymorphism of the PPARγ gene in the Danish MONICA cohort: homozygosity of the G allele confers a decreased risk of the insulin resistance syndrome. J Clin Endocrinol Metab 2002;87:3989-92.
14. Doney A, Fischer B, Frew D, Cumming A, Flavell D, World M, et al. Haplotype analysis of the PPARγ Pro12Ala and C1431T variants reveals opposing associations with body weight. BMC Genet 2002;3:21.
15. Doney ASF, Fischer B, Cecil JE, Boylan K, McGuigan FE, Ralston SH, et al. Association of the Pro12Ala and C1431T variants of PPARG and their susceptibility to type 2 diabetes. Diabetologia 2004;47:555-8.
16. Valve R, Sivenius K, Miettinen R, Pihlajamaki J, Rissanen A, Deeb SS, et al. Two polymorphisms in the peroxisome proliferator-activated receptor-γ gene are associated with severe overweight among obese women. J Clin Endocrinol Metab 1999;84:3708-12.
17. Meirhaeghe A, Cottel D, Amouyel P, Dallongeville J. Association between PPARγ haplotypes and the metabolic syndrome in French men and women. Diabetes 2005;54:3043-8.
18. Tai SE, Corella D, Deurenberg-Yap M, Adicons X, Chew SK, Tan CE, et al. Differential effects of the C1431T and Pro12Ala PPARγ gene variants on plasma lipids and diabetes risk in an Asian population. J Lipid Res 2004;45:674-85.
19. Wei Q, Jacobs DR, Schreiner PJ, Siscovick DS, Steffes MW, Fornage M. Patterns of association between PPARγ genetic variation and indices of adiposity and insulin action in African-Americans and whites: the CARDIA Study. J Mol Med 2006;84:955-65.
20. Hara M, Alcoser SZ, Qaadir A, Beiswenger KK, Cox NJ, Ehrmann DA. Insulin resistance is attenuated in women with polycystic ovary syndrome with the Pro12Ala polymorphism in the PPARγ gene. J Clin Endocrinol Metab 2002;87:772-5.
21. Kang SE, Park SY, Kim HJ, Kim SC, Ahn CW, Cha BS, et al. Effects of Pro12Ala polymorphism of peroxisome proliferator-activated receptor γ2 gene on rosiglitazone response in type 2 diabetes. Clin Pharmacol Ther 2005;78:202-8.
22. Orio F Jr, Matarese G, Di Biase S, Palomba S, Labella D, Sanna V, et al. Exon 6 and 2 peroxisome proliferator-activated receptor-γ polymorphisms in polycystic ovary syndrome. J Clin Endocrinol Metab 2003;88:5887-92.
23. Meirhaeghe A, Fajas L, Helbecque N, Cottel D, Lebel P, Dallongeville P, et al. A genetic polymorphism of the peroxisome proliferator-activated receptor γ gene influences plasma leptin levels in obese humans. Hum Mol Gen 1998;7:435-40.
24. Wang XL, Oosterhof J, Duarte N. Peroxisome proliferator-activated receptor gamma C161T polymorphism and coronary artery disease. Cardiovasc Res 1999;44:588-94.
25. Orio F Jr, Palomba S, Cascella T, Di Biase S, Labella D, Russo T, et al. Lack of an association between peroxisome proliferator-activated receptor-γ gene Pro12Ala polymorphism and adiponectin levels in the polycystic ovary syndrome. J Clin Endocrinol Metab 2004;89:5110-5.
26. Masud S, Ye S. Effect of the peroxisome proliferator activated receptor-gamma gene Pro12Ala variant on body mass index: a meta-analysis. J Med Genet 2003;40:773-80.
27. Tönjes A, Scholz M, Loeffler M, Stumvoll M. Association of Pro12Ala polymorphism in peroxisome proliferator-activated receptor gamma with pre-diabetic phenotypes: meta-analysis of 57 studies on nondiabetic individuals. Diabetes Care 2006;29:2489-97.