Introduction
The human prothrombin gene spans 21 Kb on chromosome 11p11-q12 and consists of 14 exons and 13 introns accounting for 90 percent of the sequence. A transition (G to A) has been identified at nucleotide 20210 in the 3´-untranslated region of the prothrombin gene as a risk factor for deep vein and cerebral venous thrombosis. Patients with 20210G>A mutation frequently show plasma prothrombin levels around a third higher than normal (1). The prevalence of this variant differs among populations, for example ranging from 1% to 4% reported for Caucasians (2). Recent investigations showed that variants other than 20210G>A of the prothrombin gene suggests that additional nucleotide substitutions may contribute to the development of thrombotic events and adverse pregnancy outcomes and obstetric complications (3). The 20209C>T variant of the prothrombin gene has been found in individuals of African, Caucasian and Jewish-Moroccan origins, associated with venous thrombosis, pulmonary embolism, peripheral vascular disease, deep vein thrombosis, cerebrovascular accidents (4-14) and obstetrical complications (fetal loss or intrauterine growth retardation) (3). Initial reports differ as to whether or not this prothrombin variant is associated with an increased risk of venous thromboembolism (15).
We describe the first homozygous 20209C>T mutation for a Spanish patient who developed deep vein thrombosis.
Case report
The 56-year-old male patient with a partial tear of the Achilles tendon developed calf (tibial) deep vein thrombosis after immobilization and was treated with an anticoagulant (Sintrom). The aragonese patient not seemed to have a personal history associated with thrombotic events not relevant family history. Coagulation parameters were determined later of acute thrombosis phase and the results were: free protein S 103.3% (55-150%), chromogenic protein C 102% (60-120%), antithrombin III 86% (80-120%), homocysteine 10.04 µmol/L (5-15 µmol/L), SCT-screen/confirm ratio 0.96, lupus anticoagulant screening/ confirm ratio 1.03, activated protein C resistance 2.74 ratio, factor VIII coagulant 119.6% (60-120%), and factor VII coagulant 122.9% (70-120%). Hypercoagulable studies were normal except for a slight increase in the percentage of factor VII.
Materials and methods
To determine if the deep vein thrombosis was of genetic origin, a peripheral blood DNA sample was analysed for the presence of the three most frequent mutations associated with thrombotic events: factor V Leiden (1691G>A), prothrombin (20210G>A) and methylene tetrahydrofolate reductase (MTHFR) (677C>T).
Samples from control patients (thirty patients without thrombotic problems) with the sample of index case and his first-degree relatives were analyzed in genetic department.
All described studies conducted in samples from the index case and later of his relatives were authorized by patients, who signed the corresponding informed consent. The study protocol was approved by the research ethics committee of the government of Aragón (Spain).
Genetic analysis
In the differential diagnosis of thrombotic disorders, genotyping of prothrombin 20210G>A is now performed as routine analysis. Several different molecular genetics techniques that use restriction enzymes, allele-specific amplification, hybridization probes at a single detection temperature or the LightCycler system have been described (8,16-19).
In our laboratory we used PTH-FV-MTHFR test StripAssay (Vienna Lab) (Izasa, Barcelona, Spain) based on the Polymerase Chain Reaction (PCR) and reverse hybridization/revealed using allele specific probes. The patient’s DNA extracted from EDTA Blood was analysed initially by method PTH-FV-MTHFR test Strip Assay and it was found that it could not be determined the presence or absence of the normal allele of prothrombin by the method described.Genomic DNA was extracted from peripheral blood by standard procedures (EZ1DNA blood 350 µl Kit using magnetic particle technology in EZ1 Advanced (1–6 samples per run) instrument).
The PTH-FV-MTHFR test StripAssay is a test designed to detect mutations in factor V (FV), prothrombin (PTH) and MTHFR gene based PCR and hybridization reverse. The procedure consists of three steps: a) isolation of DNA; b) amplification by PCR using primers labeled with biotin; c) reverse hybridization/detection of amplification products in a strip containing oligonucleotide probes allele-specific fixed in parallel lines. The sequences labeled with biotin attached to the strip are detected using streptavidin-alkaline phosphatase and color substrate.
The sample of the patient was again analyzed two times more with new venous extraction and the same result was obtained. It was not possible to determine the presence or absence of the normal allele of phrotrombin gene.
The blood sample was analyzed with The Xpert® HemosIL® Factor II & Factor V Assay (Cepheid/Instrumentation Laboratory), which is a qualitative in vitro diagnostic genotyping test for the detection of Factor II (20210G>A mutation) and Factor V (1691G>A mutation) alleles from sodium citrate or EDTA anticoagulated whole blood. The GeneXpert Dx System automates and integrates sample purification, nucleic acid amplification, and detection of the target sequence in whole blood using real-time PCR assays.
Real time-PCR,Quantitative PCR, was performed with the LightCycler System 2.0 (Roche Diagnostics, Manheim, Germany) using the capillaries of 20 μL. We used the LightCycler FastStart DNA Master Plus SYBR GreenI.The hot start effect of FastStart Taq DNA polymerase minimizes the formation of non-specific products and improves sensitivity for the desired target. Also, we used Uracil-DNA Glycosylase, heat-labile (Roche Diagnostics) 1U for to eliminate PCR carry over contaminations.Fluorescence protocols PCR was performed with 0.2 Μm primers in a standard PCR reaction, supplemented.Samples were spun into glass capillary cuvettes, capped and placed in the Light-Cycler. For prothrombin gene mutation, amplification was performed for 40 cycles of denaturation (94 °C, 0 s, ramp rate 20 °C/s), annealing (50 °C, 10 s, ramp rate 20 °C/s), and extension (72 °C, 10 s, ramp rate 2 °C/s). After amplification, a melting curve was generated by holding the reaction at 50 °C for 30 s, and then heating slowly to 94 °C with a ramp rate of 0.2 °C per s. Fluorescence was collected continuously during the slow temperature ramp. Melting curves were converted to melting peaks by plotting the negative derivative of the fluorescence with respect to temperature (-dF/dT) against temperature. The entire process takes approximately 30 min with no separate product manipulation necessary.A single fluorescence reading for each sample was taken at the annealing step. At this stage of the PCR cycle both probes are allowed to hybridize and the total amount of product formed was determined by the log-linear slope intercept of the X-axis.
In order to confirm the findings by LightCycler analysis it was done Sanger sequencing. The sequence of the prothrombin gene was amplified by PCR using total DNA from whole blood as template. The amplified products were purified and directly sequenced using an automated DNA sequencer (capillary sequencing) (model 3130, Applied Biosystems, Foster City, CA).
Prothrombin gene was individually amplified by PCR using genomic DNA as template. Sequences of oligonucleotides used for amplification and sequencing were deduced from the published Human Genome sequence (
http://genome.ucsc.edu). Genbank Prothrombin cDNA entry with accession number NM_000506 was used as a reference sequence for analysis of mutations and numbering of nucleotides. Codon numbering is given for mature prothrombin protein. To identify the exact nucleotide change (the wild-type C was replaced by a T at nucleotide position 20209), we designed PCR and sequencing primers (5´-CCGCCTGAAGAAGTG GATA-3´ and 5´-CAGAGAGCTGCCCATGAAT-3´ for the forward and reverse direction, respectively).
Results
The results of mutations studied obtained for the patient and his first-degree family members are presented in summary form in Table 1. The patient was heterozygous for 677C>T and normal for 1691G>A (Figure 1). The fact that it was not possible to determine the presence or absence of the normal allele for factor II using the FV-PTH-MTHFR StripAssay or the Xpert® HemosIL® Factor II & Factor V Assay was suggestive that the patient had a variant interfering with the PCR product. As can be observed from Figure 1, it was not possible to determine the presence or absence of the normal allele for prothrombin in the sample of index case. In the strips for subjects with the homozygous 20209C>T mutation, no blue band was detected (wild-type 20210G>A), and in the heterozygous 20209C>T mutation, only a partial blue band was detected (very little blue). Neither of the two methods confirmed the presence or absence of the normal allele for prothrombin gene (Figure 1).
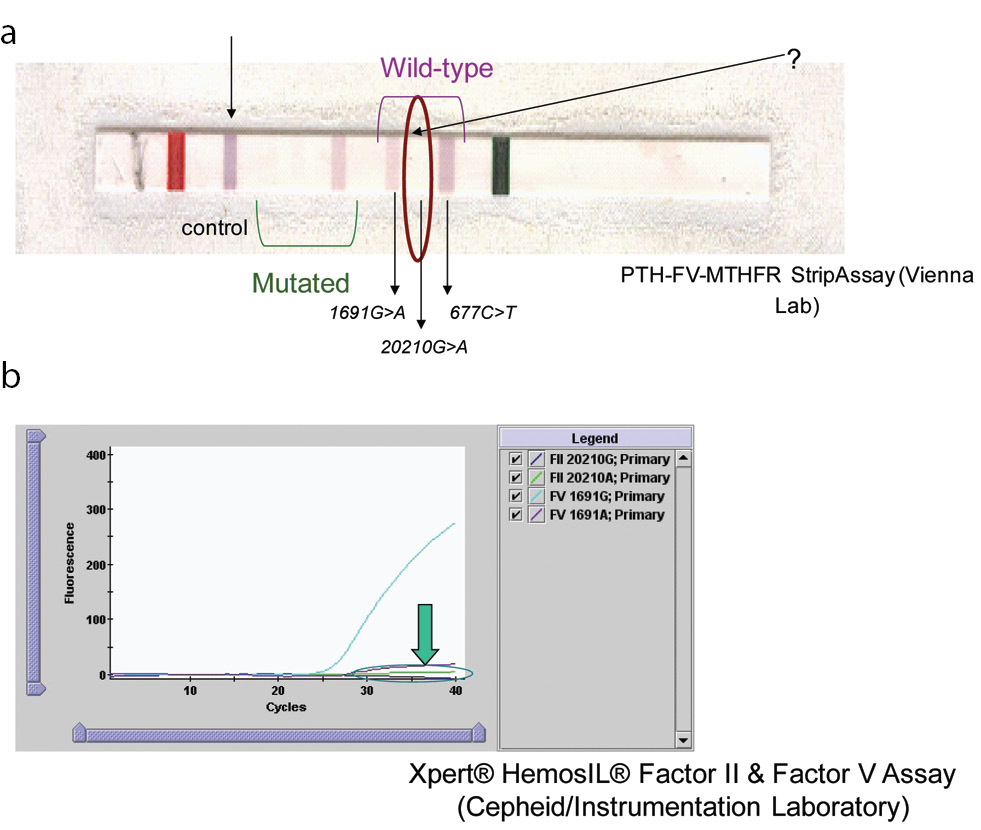
Figure 1. Results for index patient using two different methods.
(A). Results for patient blood sample analysed by the PTH-FV-MTHFR StripAssay (ViennaLab) method. In 20209C>T homozygous cases, no blue band is observed in the strip. (B). Result obtained with the Xpert® HemosIL® Factor II & Factor V Assay.
The presence of this 20209C>T mutation was confirmed in another DNA sample from the patient and DNA samples from first-degree family members were also studied to determine its presence or absence. Heterozygous 20209C>T was detected in the patient’s parents and brother, homozygous 20209C>T was detected in the patient’s sister and heterozygous 20209C>T status was confirmed for the patient’s son and daughter (Table 1).
Table 1. Status of factor II (20209C>T and 20210G>A), factor V Leiden (1691G>A) and MTHFR (677C>T) mutations in the index patient and in first-degree family members.
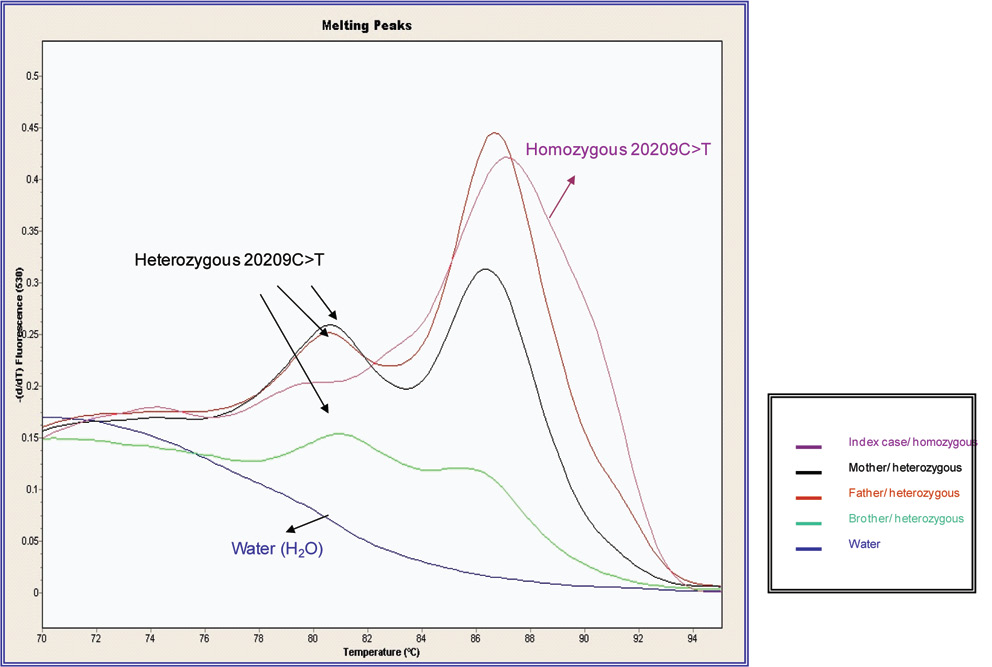
Figure 2 shows the results by Real time-PCR method (Light-Cycler) for the samples analysed for the index case, his parents and brother, indicating the patient’s homozygosis status. Figure 3 shows the direct DNA sequencing for the patient and his parents.
Figure 2. DNA melting curves for the patient and first-degree relatives obtained using LightCycler. The melting curve of the heterozygous for 20209 C>T demonstrates peaks at 87 ºC and 81 ºC. Patient homozygous for 20209C>T presents one peak at 87ºC.
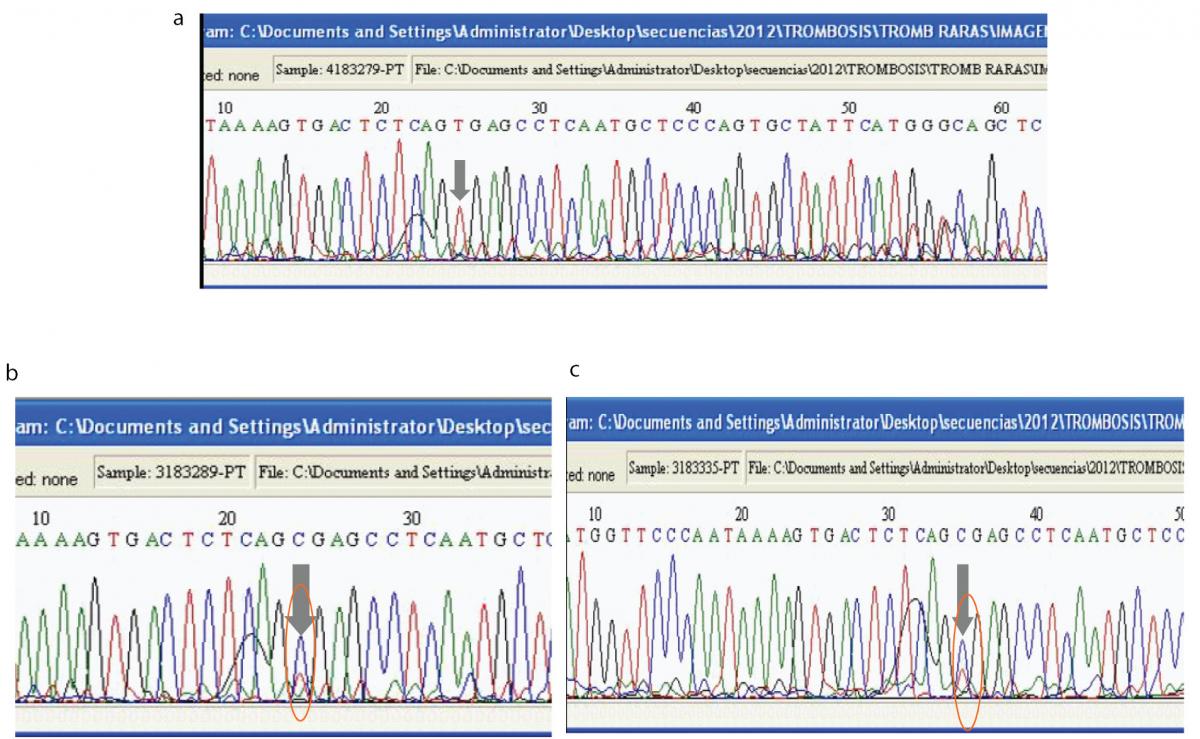
Figure 3. Direct DNA sequence analysis with designed primers to identify the 20209C>T mutation in the index case.
(A). Sequence analysis for the patient in the forward direction shows only one peak (red) at prothrombin nucleotide position 20209. The grey arrow shows that the DNA is homozygous for a C to T mutation at position 20209. (B). Sequence analysis for the patient’s father in the forward direction shows two overlapping peaks (the blue peak corresponds to C and the red peak corresponds to T) at prothrombin nucleotide position 20209, which means that this individual is heterozygous for the 20209C>T mutation. (C). Sequence analysis for the patient’s mother in the forward direction shows two overlapping peaks (the blue peak corresponds to C and the red peak corresponds to T) at prothrombin nucleotide position 20209, which means that this individual is heterozygous for the 20209C>T mutation.
Thirty control patients samples (patients without thrombotic problems) were analyzed and did not show the mutation 20209C>T heterozygous or homozygous status.
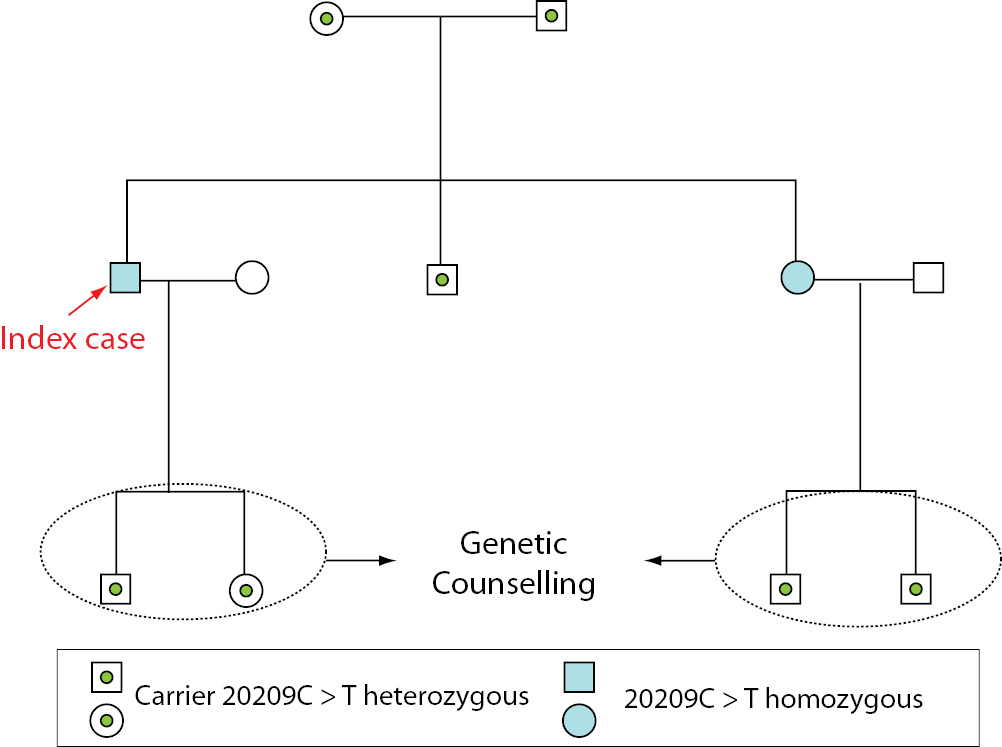
Because they are from the same village, the parents are probably consanguineous. The patient and his brother and sister are also 677C>T heterozygous. The homozygous presence of the 677C>T mutation, as well as being heterozygous for 20209C>T, in the father of the index case may explain his clinical signs: thrombophlebitis. The presence of homozygous 20209C>T was the probable cause of the patient’s deep vein thrombosis and the association with heterozygous 677C>T may increase the risk of thrombotic events. The patient’s sister has the same mutations as the index case and has got some symptoms (frequent bruising, varicose veins), so that knowing the presence of this mutation will help prevent thrombotic events in this patient. The two subjects with homozygous 20209C>T mutations have children and so will need genetic counselling (see pedigree, Figure 4).
Figure 4. Pedigree with the results for 20209C>T for family members.
Discussion
We found that the patient had a homozygous C>T mutation at position 20209; this interfered with the PCR product, which needs a C at this position to be able to bind to the wild-type probe present in the test strip (Vienna Lab method). If this patient had not been studied using the Vienna Lab method, the presence of 20209 C>T would not have been detected.
The 20209C>T mutation in samples gave unusual genotyping results in laboratories that participate in external quality assurance (EQA) programmes (20). The fact that the pyrosequencing method was not able to detect the 20209C>T mutation confirms other reports that some methods are unable to detect the presence of this mutation (19).
We have no found presence of 20209C>T variant in our control group (thirty samples). In the literature has been described the incidence of the prothrombin 20209C>T variant in a case control study of risk factors for venous thromboembolism in African-American and Caucasian populations. The incidence in African-Americans was 0.7% in controls versus 1.3% in cases and in the Caucasian population, one patient was found to be heterozygous in the control group and none in the study group (9). Another study of a black population with thrombosis reported incidences of 4.9% (4/81) (11) and of 1.6% (1/61) (21). Patients with heterozygous prothrombin 20209C>T present a risk factor for venous thromboembolism. Hereditary thrombophilia has autosomal dominant inheritance pattern and is associated with an increased risk of venous thrombosis: deficiency of antithrombin III, protein C, or free protein S, or factor V Leiden, or mutations in phrotrombin gene. The classic prothrombin mutation associated with an increased thrombotic risk is a G to A substitution at position 20210 of the prothrombin gene. The 20209C>T mutation may also be associated with thrombosis (9,22).
Some studies reported that elevated plasma prothrombin activity not appeared in patients with 20209C>T mutations, whereas recent studies have shown that this mutation is associated with gain-of-function of 3´end processing and upregulation of prothrombin protein expression (15). The case index, homozygous for the 20209C> T mutation, had not elevated levels of prothrombin which was in agreement with that described in the literature. Likewise, the sister of the index case homozygous for the 20209C> T mutation and the other studied first-degree relatives who were heterozygous for this mutation showed no elevated levels of prothrombin or alteration in any of the hypercoagulable study parameters. Another study showed a contradictory outcome for in vitro functional studies of the 20209C>T variant (12).
The patient’s parents were both 20209C>T heterozygous and this mutation could explain the mother’s acute myocardial infarction and the father’s thrombophlebitis (20209C>T heterozygous plus 677C>T homozygous). The patient’s brother was 20209C>T heterozygous and asymptomatic and the patient’s sister was 20209C>T homozygous and symptomatic (bruising and varicose veins).
Conclusions
In the index case described in this work could justify the phenomenon of deep vein thrombosis by the presence of the mutation 20209C>T in homozygosity and also in the presence of the mutation 677C>T in heterozygosity because the combination of mutations would increase the risk of thrombosis.
Larger and population-specific studies of Spanish individuals with thrombotic (especially patients with deep vein thrombosis and with screening of the three common mutations associated with thrombosis negative) and/or obstetric complications will be necessary to establish the true prevalence of the 20209C>T mutation and evaluate the thrombotic and/or obstetric consequences. The presence of the mutation 20209C>T both homozygous and heterozygous in combination with the presence of the mutation 677C>T heterozygous or homozygous might be a risk factor to consider in the face of suffering a possible thrombotic event. Suitable genetic counselling should be provided to the patient and first-degree relatives (siblings and offspring of childbearing age) as it important to detect prothrombin gene variants that could increase their risk for thrombotic events.
The possibility of including 20209C>T mutation detection in screening for the most common mutations associated with thrombotic events should be considered.
Acknowledgements
We thank the Department of Genetics of Hospital Universitario Miguel Servet in Zaragoza for its collaboration with this research. Writing assistance: Ailish MJ Maher provided English language editing.