Introduction
Proteomics is the study of proteins expressed by an organism or cellular system (proteome), in order to understand cellular processes. This is an area that has been widely explored in the past decade and during this time highly sensitive techniques have been developed for analyzing the proteome of biological samples such as cells, tissues and body fluids (1,2). Progress that has been made in new technologies, has led to the discovery of many potential biomarkers. These molecules are measurable indicators of a biological state (3), be it normal or abnormal, and therefore can be used in a clinical setting in disease diagnosis and management.
Biomarkers can be of varied chemical nature, be they genetic (DNA and RNA), proteins, secondary metabolites or carbohydrates. Although genetic and protein biomarkers are the most studied, the latter have gained recognition due to the close correlation that exists between the proteome and the biological activity of the cell or system. Proteins are directly responsible for cell function, thus abnormal protein expression is an indication of cellular disruption due to a pathological condition (4).
Biomarkers can be found in different biological samples. Blood has been the most widely studied source of biomarkers to date, as it is in contact with all the cells in the body. However, proteomic analysis of blood samples poses certain disadvantages. During sample collection proteases are often activated, which generates a range of proteolytic products, thus introducing variability to the sample (5). Furthermore, blood contains 20 high abundance proteins which correspond to 99% of the proteins in the sample (6,7); these high abundance proteins mask other less abundant, potentially interesting proteins. Due to this, urine has roused great interest in recent years. Urine is an ultra filtration of the blood in the body and is very stable compared to blood (8,9). It is easy to process in the lab, large amounts can be obtained, and its collection is simple and non-invasive causing minimal stress to patients (10). The impact of urinary biomarkers in a clinical setting lies in these advantages, as they could provide valuable information without the need for costly and invasive screening/diagnostic/monitoring procedures for the patient. This could also translate into improved, early screening programs, in order to reduce illness and death, thus diminishing the burden on the healthcare system. The main disadvantage of urine is the variation in protein concentration due to differences in fluid consumption during the day; however, this can be countered by normalizing with creatinine (7).
The Human Proteome Organization (HUPO) approved the Human Kidney and Urine Proteome Project in October 2007 as one of the HUPO-sponsored scientific initiatives, in which more than 150 participating scientists collect and publicly share proteomics data on urine. Approximately 1500 proteins have been shown to constitute the urinary proteome (11), of which large proportions are extra cellular proteins, plasma membrane proteins, and lysosomal proteins. Several studies have indicated that under physiological conditions approximately 30% of the proteins in normal human urine are plasma proteins, while the other 70% are proteins derived from the kidney and genitourinary tract (12-15). Therefore, as expected, urine is an interesting source of biomarkers for monitoring the health of kidneys and genitourinary organs (such as the prostate), which secrete proteins into the urine (16). However, protein biomarkers have also been identified in urine for other types of disorders, such as different types of cancer, metabolic diseases, and even immunological disorders (9,17,18).
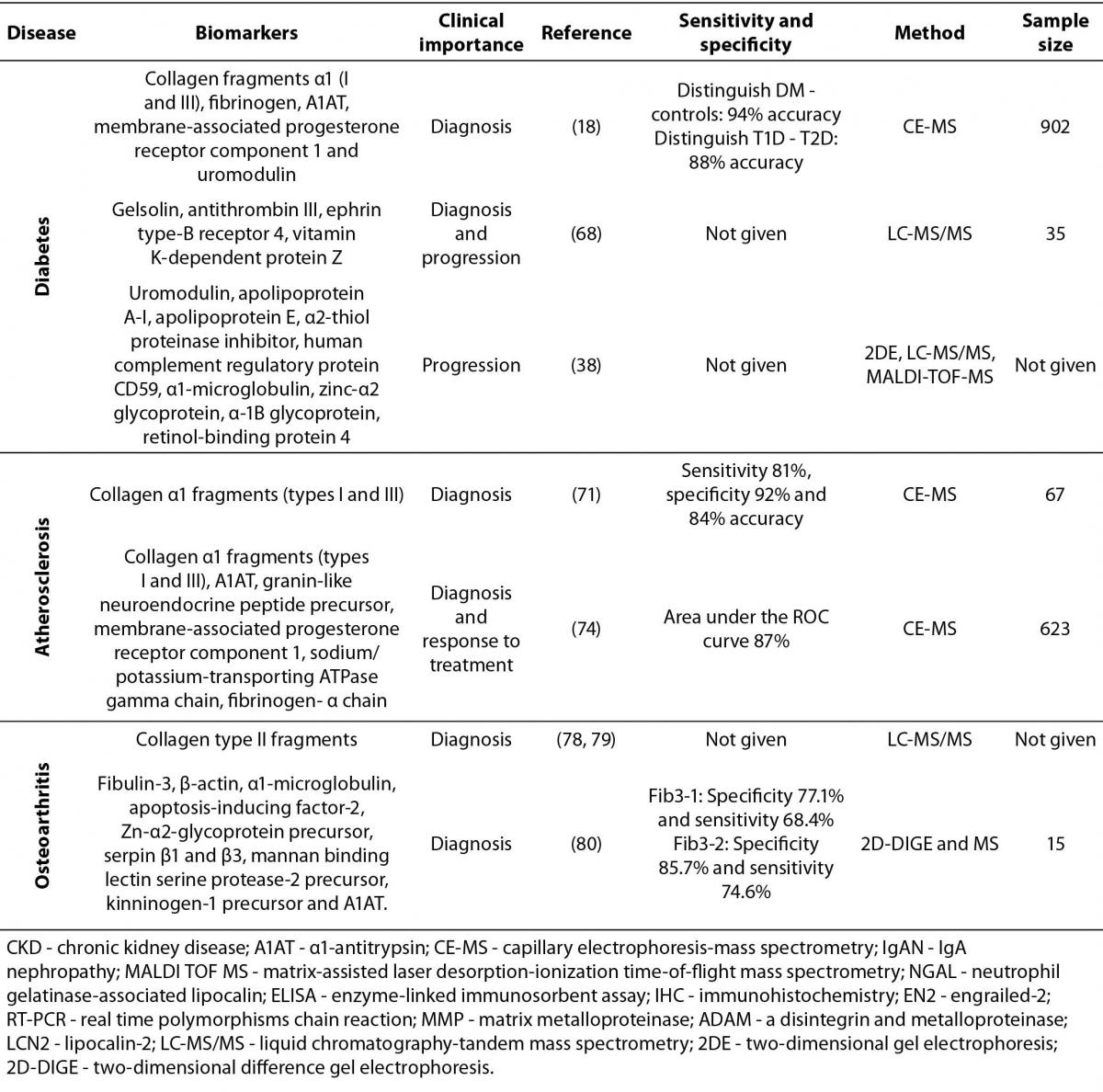
A search of the literature reveals many biomarker candidates identified in urine by different strategies; however, few have been implemented in a clinical setting. One of the biggest challenges is that biomarkers are often shared by several pathologies so are not specific to one disease, as evidenced in this review. Rather than a single urinary protein biomarker, a panel of biomarkers may be required to achieve the overall high level of specificity needed. When seemingly contradictory results are seen in this light, they may be understood as complementary. The aim of this review is to present an overview of some of the recent protein biomarkers found in urine, and their potential use in the diagnosis of different pathologies involving the kidney and urinary tract, as well as of cancer, metabolic and inflammatory diseases. Integrating information from different studies will allow for disease-specificity of biomarker candidates to be better evaluated, by comparing studies across various diseases. In the first part of this review, we summarize a few of the potential protein biomarkers recently detected in urine for chronic kidney disease, glomerulonephritis and kidney transplant rejection. This is followed by advances in urinary protein biomarkers for prostate and breast cancer, as well as diabetes, atherosclerosis and osteoarthritis. In addition, we summarized the biomarkers described for each condition, including clinical importance of the biomarkers, references of main studies, sensitivity and specificity, types of methods used in the studies and sample size (table 1).
Table 1. Summary of biomarkers described in this review.
Materials and methods
The keywords “urine, protein, biomarkers” used in a general PubMed search yielded over 10,000 papers on the subject. Due to the wide range of publications available, it was necessary to concentrate on a few important diseases for which urinary protein biomarkers have been proposed. We selected a couple of diseases affecting the urogenital tract, specifically chronic kidney disease and prostate cancer, as well as a few non-urogenital pathologies, such as breast cancer, diabetes, atherosclerosis and osteoarthritis. A more limited PubMed search using keywords “urine, biomarker, protein, and/or prostate cancer / breast cancer / chronic kidney disease / diabetes / atherosclerosis / osteoarthritis “, yielded 2,349 papers overall. The search was limited to papers published in English in the past 10 years, with abstract and full text available. The titles and abstracts were reviewed and the most relevant publications were selected. Due to the broad character of the review subject, we focused mainly on novel studies with larger human cohorts.
Kidney disease
Chronic kidney disease (CKD) is characterized by the progressive loss of renal function. This disease may take decades to progress and can eventually lead to kidney failure, requiring dialysis or a kidney transplant for survival (19). The initial diagnostic tests for CKD include measuring proteinuria and serum creatinine; however, it has been shown that these diagnostic aids are not sensitive enough in the early stages of the disease or specific to the variations between individuals (19-21).
Many potential protein biomarkers for CKD have been suggested, including a panel of 273 CKD specific peptides reported by Good and colleagues (19) in 2010 for the early diagnosis of the disease. In this study urine samples were collected from 3,600 individuals in over 20 clinical centres around America, Europe and Australia, and were analyzed using capillary electrophoresis-mass spectrometry (CE-MS). This biomarker pattern of 273 peptides was able to distinguish between healthy individuals and those with CKD with a specificity of 100% and a sensitivity value of 98.7%. This model was then validated in an independent blinded cohort of 144 individuals with sensitivity results of 85.5% and 100% specificity. These proteins differentially expressed in urine of patients with CKD included different collagen fragments, serum proteins like α1-antitrypsin (A1AT), albumin, haemoglobin α chain, and fibrinogen α chain, as well as kidney-specific proteins such as uromodulin, sodium/potassium-transporting ATPase γ chain, and membrane-associated progesterone receptor component 1. Despite the high specificity achieved it must be noted that the training set used in this study included patients with various biopsy-proven kidney diseases, such as IgA nephropathy, diabetic nephropathy, vasculitis, lupus and glomerulonephritis; so this panel of biomarkers is not specific for one disease, but for kidney disease in general.
IgA nephropathy
Several conditions have been associated with the onset of CKD, including diabetic nephropathy, hypertension and glomerulonephritis (22). Kidney damage observed in glomerulonephritis is characterized by changes in glomerular permeability and structure. Selective filtration mainly occurs in the glomerular basement membrane where proteins are excluded based on their size and charge. However, in glomerulonephritis permeability is altered leading to serum proteins leaking into urine. Different types of glomerulonephritis have been clinically described, the most common type being IgA nephropathy (IgAN). IgAN leads to end stage renal disease within 25 years of the manifestation of the disease, in approximately 25% to 30% of cases (23). The current clinical practice for confirming IgAN diagnosis is renal biopsy. This has driven researchers around the world to work on developing non-invasive diagnostic tools for the early detection of IgAN.
In a study published in 2005 (24), urine samples from 45 patients with IgAN, 13 patients with membranous nephropathy and 57 healthy controls were screened using CE-MS. This study revealed that the urinary proteome of patients with IgAN was significantly different and could be successfully used to distinguish those patients with IgAN from the other two study groups, even when protein excretion into urine was within the normal range. Another interesting finding was that as patients with IgAN underwent treatment their urinary proteome changed adopting a pattern similar to that displayed by the control subjects. From this study we are able to conclude that IgAN patients exhibit a specific urinary proteomic profile that can differentiate them from healthy individuals and from patients with other renal diseases, as well as allowing for treatment effectiveness to be monitored; however, the identity of the peptides in this panel of biomarkers was not established.
In a recent study (25), 55 peptides that differentiated between IgAN patients and healthy subjects were isolated from blood and urine samples taken from 33 individuals (19 with biopsy-proven IgAN and 14 controls). Sensitivity and specificity data were not given. Sixteen of these 55 peptides were identified in urine and corresponded to uromodulin, α1-antitrypsin and β2-microglobulin peptides. These peptides were also correlated with poor histological lesions typical of patients with IgAN, particularly tubulointerstitial damage and segmental glomerulosclerosis. However, it seems that the increased expression of A1AT is due mostly to the inflammatory process rather than being a specific response to the disease, so is therefore not a specific biomarker for IgAN. Similarly, uromodulin has been implicated in other renal diseases so is also not a specific biomarker for IgAN (26,27).
Tubulointerstitial injury plays an important role in the development of IgAN and is typically diagnosed by measuring the levels of N-acetyl-β-D-glucosaminidase (NAG) in urine or in tissue. However, NAG is a lysosomal enzyme that is increased when there is damage to the renal tubules, so it can also be found at higher levels in patients suffering from diseases such as nephrotic syndrome, glomerulonephritis and diabetic nephropathy. Since NAG is not a specific biomarker of IgAN it was necessary to search for more specific biomarkers of the disease and neutrophil gelatinase-associated lipocalin (NGAL) was suggested. Ding and colleagues (28) found that urinary levels of NGAL were significantly higher in patients with IgAN compared to healthy controls, even when increased NAG levels could not yet be detected. NGAL is more specific (67.9%) and sensitive (90.2%) for IgAN detection compared to NAG (82.4% sensitivity and 60.5% specificity) suggesting that urinary NGAL is a superior biomarker of early tubulointerstitial injury in IgAN. However, this corresponds to a preliminary study with 70 patients and needs to be validated with a larger cohort of patients.
Cancer
Cancer is a serious public health concern worldwide. Each year more than 12 million people are diagnosed with some type of cancer and approximately 7 million people die from cancer each year (29). The highly heterogeneous nature of cancer provides a real challenge for clinical disease management; clinical progression is often difficult to predict and treatment is therefore not as effective as it should be. Consequently, the use of biomarkers as an aid for early diagnosis and monitoring progression and treatment effectiveness has been extensively studied.
Prostate cancer
Prostate cancer is the second most prevalent cancer in men worldwide after lung cancer. The International Agency for Research on Cancer in its latest report (29) estimated a global incidence of 899,000 new cases of prostate cancer per year and an annual mortality rate of 258,000 men due to this disease.
The classic diagnostic tools for prostate cancer detection are the measurement of prostate specific antigen (PSA) in serum, digital rectal examination and imaging techniques. The least invasive of these is the measurement of PSA levels in the blood, which is a protein naturally produced by the prostate that helps keep the semen in its liquid form. PSA levels are elevated as a result of prostate cancer; however, there are other factors such as benign prostatic hyperplasia, age and sexual activity that also raise PSA levels, so this test produces many false positives (30). There are also some prostate cancers that exhibit normal PSA levels and therefore are not detected in time (31), which leads to more aggressive treatment later on. In the last few years there has been great progress into alternate ways of using the PSA test with better performance. The Prostate Health Index (PHI) is a new formula that combines all three forms of PSA (total PSA, free PSA and proPSA) into a single score that can be used to aid in diagnosis and management. A recent study has shown (32) that proPSA and PHI can discriminate between patients with prostate cancer and those with benign prostatic hyperplasia or chronic histological prostatic inflammation, thus reducing the need for unnecessary biopsies by 27%. However, PHI is not effective for stratification of patients.
Advances are also being made on potential urinary biomarkers for prostate cancer. Annexin A3 is a calcium-binding protein that plays a role in the regulation of cellular growth and in signal transduction pathways, and as such has been shown to have important roles in tumour development, metastasis and drug resistance (33). A large study using urine samples from 591 patients (34) reported Annexin A3 as a novel urine-based biomarker for early prostate cancer detection when used in conjunction with PSA. The accuracy of the combined annexin A3/PSA test was superior to PSA alone (area under the ROC curve of 0.82 versus 0.68).
Using MALDI-TOF (matrix-assisted laser desorption-ionization time-of-flight) another large study (35) of 407 urine samples identified uromodulin and semenogelin as protein biomarkers of interest that could distinguish between prostate cancer and benign prostatic hyperplasia with 71.2% sensitivity and 67.4% specificity. Semenogelin, a seminal vesicle derived protein found in seminal fluid, was over expressed in patients with prostate cancer; while uromodulin, an abundant protein in normal human urine, was down regulated in these patients. Despite the large dataset in this study, the sensitivity and specificity of these proteins was quite low, hence they are not prime candidates to be used on their own. Changes in uromodulin have also been detected in diabetes (18,36) and chronic kidney disease (19); and it has been accepted by the US Food and Drug Administration, European Medicines Agency, and Pharmaceuticals and Medical Devices Agency, as a urinary biomarker for detecting drug-induced kidney injury during preclinical toxicological testing (37).
In 2008, Theodorescu and colleagues (9) used CE-MS to identify and validate 12 biomarkers from first void urine that were able to identify patients with prostate cancer with 91% sensitivity and 69% specificity. The identified biomarkers were sodium/potassium-transporting ATPase γ, collagen α-1 (III), collagen α-1(I), psoriasis susceptibility 1 candidate gene 2 protein, hepatocellular carcinoma associated protein TB6, histone H2B, osteopontin, polymeric Ig receptor, transmembrane secretory component, prostatic acid phosphatase, fibrinogen α chain precursor, and semenogelin 1, most of which were found to be down-regulated in patients with prostate cancer. This study supports the use of a panel of biomarkers for disease diagnosis rather than a stand-alone biomarker.
Recently, engrailed-2 (EN2), a homeobox protein involved in the development of the normal prostate gland, was found to detect prostate cancer in urine with 66.7% sensitivity and 89.3% specificity using an ELISA-based assay (38,39). This protein was found in urine of prostate cancer patients but was no longer detectable after prostatectomy in the same patients, nor in healthy controls, indicating a diagnostic potential for EN2. The high predictive value of urinary EN2 raises the possibility that it could be used alongside PSA in the diagnosis of prostate cancer, in order to increase sensitivity and specificity in diagnosis, thus picking up those cancers that would go undetected, and also reducing the number of false positives thus avoiding unnecessary prostate biopsies. In a later study by the same research group (40), a positive correlation was shown between urinary levels of EN2 and prostate cancer volume, marking it as a useful biomarker not only for prostate cancer diagnosis but also for risk stratification. To date this is the first biomarker of prostate cancer that correlates with the amount of cancer present, and could potentially aid in making clinical decisions regarding disease management (treatment vs. surveillance). Currently none of these urinary protein biomarkers have been introduced into clinical practice.
Breast cancer
Prostate cancer is a urogenital disease, so it is logical to think that biomarkers for this disease can easily be found in urine, however other non-urogenital cancers also exhibit changes in the urinary proteomic profile; an example of this is breast cancer. Breast cancer is by far the most frequent cancer among women, with an estimated 1.38 million new cancer cases per year and an annual mortality rate of 458,000 women worldwide (29). If breast cancer is detected during its earlier stages, the 5-year survival rate may be as high as 93% (41), making early detection essential for favourable prognosis. Current guidelines for the early detection of breast cancer include mammography and clinical breast examination. However, not all cases of breast cancer are detected by a mammogram, and many women experience false alarms. Results from several randomized screening trials (42,43) suggest that mammography reduces breast cancer mortality rate by 15% to 20%. Tumour markers currently in use in the evaluation of breast cancer, as recommended by the National Academy of Clinical Biochemistry (NACB) (44), include estrogen and progesterone receptors, human epidermal growth factor receptor-2, urokinase plasminogen activator/plasminogen activator inhibitor 1 and cancer antigen 15-3. These are useful for predicting response to therapy, but show little potential for early detection (45). In recent years, several studies have reported urinary biomarkers for breast cancer detection (46); among those reported are the matrix metalloproteinase’s (MMPs) and ADAMS (A Disintegrin And Metalloproteinase). These proteins are zinc proteases that degrade the components of the extra cellular matrix, and as such are involved in the process of tumour progression, from angiogenesis and cell migration, to remodelling of the tumour microenvironment, invasion, and metastasis (47,48).
A study of 148 women (49) showed that women with high levels of MMP-9 and ADAM-12 in urine were 5 times more likely to develop atypical hyperplasia and 13 times more likely to develop lobular carcinoma in situ, which are precursors of cancer. Therefore, it might be concluded that these biomarkers not only act as indicators of the presence of breast cancer, but also as high-risk predictors. It is reported that ADAM-12, expressed by cancer cells, accelerates breast cancer progression by inducing apoptosis of stromal cells (50) and by degrading several extra cellular matrix components such as collagen type IV and fibronectin (51).
In contrast, a recent study (52) of 50 breast cancer patients and 46 matched controls did not find a significant difference in the urinary ADAM-12 levels between these two study groups, but did report an increase following surgery (P < 0.001). This could mean that the increase in ADAM-12 is due to trauma and inflammation in the tissue, rather than specifically due to breast cancer. This is supported by other studies that have reported ADAM-12 over expression in other cancer types, such as brain, colon, gastric and lung cancer (53-55).
Another MMP related protein of interest in breast cancer is NGAL. NGAL is part of the matrix metalloproteinase-9 (MMP-9) complex and has a role in protecting MMP-9 from auto degradation, thereby protecting its enzymatic activity (56). A study of 20 urine samples (57) showed that NGAL levels were significantly higher in samples from metastatic breast cancer patients compared with healthy women. These results were also consistent with immunohistochemistry results of breast tissue as well as in vitro and animal studies, leading to the conclusion that NGAL levels are associated with poor prognosis, high proliferation and decreased survival. Recent evidence suggests, however, that increased expression of NGAL may also occur in other cancer types, such as bladder, colorectal, liver, lung, ovarian, and pancreatic cancer (58), as well as in renal failure (59).
Despite the number of published studies on breast cancer biomarkers, yet there is no validated urinary biomarker available for use in routine clinical practice, therefore breast cancer detection and monitoring remains dependent mostly on invasive procedures.
Metabolic and inflammatory diseases
The search for biomarkers in urine has also been extended to metabolic and inflammatory diseases, which have high morbidity rates, such as diabetes, atherosclerosis and osteoarthritis.
Diabetes
Diabetes mellitus (DM) is a complex disease characterized by the impaired metabolism of glucose, its main symptom being the increase in blood glucose levels (hyperglycaemia). With time, these alterations cause secondary cellular dysfunctions and vascular damage including diabetic nephropathy. The disease has been classified into two main types. Type 1 diabetes (T1D) is associated with destruction of insulin-producing β-cells in the pancreas, typically by an autoimmune mechanism, leading to insufficient insulin production. Type 2 diabetes (T2D) on the other hand, is caused by insulin resistance, which is the inability of cells to respond adequately to insulin, and is often associated with obesity.
Both types of diabetes are diagnosed and monitored by measuring fasting plasma glucose concentration, haemoglobin A1C levels and with the oral glucose tolerance test (60). According to the International Diabetes Federation (61), T2D frequently goes undetected leading to further complications. In countries such as the US approximately 30% of cases are not diagnosed, while in African countries this number can be as high as 90%. For those that are diagnosed, on average a diagnosis is reached between 4 and 7 years after the onset of the disease (62). For this reason, different research groups have focused on finding early onset biomarkers.
An early study (63) evaluated the urinary proteome of 305 individuals, using CE-MS, and identified a panel of 261 biomarkers which could distinguish between diabetic patients and healthy controls with 89% sensitivity and 91% specificity. These results were later validated in a larger study (18) of 587 diabetic patients (299 T1D and 288 T2D) and 315 healthy individuals, with similar sensitivity and specificity results. The differences in T1D and T2D urine samples were also evaluated and a panel of 131 potential biomarkers was proposed. The majority of the identified biomarkers were fragments of collagen α-1 (I and III), fibrinogen, α1-antitrypsin (A1AT), membrane-associated progesterone receptor component 1 and uromodulin. One of the most predominant changes in the urinary proteome of the diabetic patients was a significant reduction in collagen fragments compared to controls. This shift was more noticeable in patients with T2D compared to those with T1D, indicating differences in extra cellular matrix remodelling. Another interesting find was that although A1AT, an acute phase protein and physiological inhibitor of serine proteases, was detected in low levels in plasma samples of diabetic patients, in urine it was significantly increased, which suggests a possible increase in degradation and clearance in the kidneys (18,64). The increased degradation of A1AT facilitates the conversion of fibrinogen to fibrin by thrombin, leading to a higher risk of atherothrombotic disorders in patients with diabetes (65).
A study conducted in 2012 (66) with 15 T1D patients, with and without complications, such as retinopathy and nephropathy, reported gelsolin and antithrombin III as promising urinary biomarkers for the early diagnosis of T1D; while ephrin type-B receptor 4 and vitamin K-dependent protein Z could be used to predict T1D progression (retinopathy and nephropathy). These proteins are associated to micro vascular complications that are so common with T1D. The limitation of this study however, is the small dataset used; so prior to drawing any conclusions it is necessary to validate these results in a larger dataset.
Another study (36) identified urinary proteins correlated with the progression and complications of T1D. Samples were taken from diabetic patients classified in two groups according to their albuminuria levels (norm-albuminuria and micro-albuminuria) and compared with healthy individuals using high-resolution two-dimensional gel electrophoresis (2DE), MALDI-TOF-MS and liquid chromatography– tandem mass spectrometry (LC-MS/MS) analysis. Uromodulin, apolipoprotein A-I, apolipoprotein E, α2-thiol proteinase inhibitor, and human complement regulatory protein CD59 were found in lower levels in diabetic patients with micro-albuminuria, while proteins such as α1-microglobulin, zinc-α2 glycoprotein, α-1B glycoprotein, and retinol-binding protein 4 were highly expressed in the same study group. These proteins are associated with microalbuminuria and therefore could be useful for monitoring the progression of T1D.
Changes in the above biomarkers are most likely due to renal complications rather than to diabetes specifically. This is supported by studies which have shown that urinary collagen and fibrinogen fragments, A1AT, uromodulin, and membrane-associated progesterone receptor component 1 are also observed in kidney disease (19). Gelsolin (67), A1AT and apolipoprotein E (68) have been detected in urine samples of patients with bladder cancer; and uromodulin (also known as Tamm-Horsfall urinary glycoprotein) has been accepted by the US Food and Drug Administration, European Medicines Agency, and Pharmaceuticals and Medical Devices Agency, as a urinary biomarker for detecting drug-induced kidney injury during preclinical toxicological testing (37).
Atherosclerosis
Atherosclerosis is a chronic, inflammatory disease of the arterial wall that underlies many of the common causes of cardiovascular morbidity and mortality, including myocardial infarction, stroke and peripheral vascular disease. Some people who have atherosclerosis have no signs or symptoms. They may not be diagnosed until after a heart attack or stroke, therefore early detection is crucial.
Several studies have evaluated biomarkers for the early detection of atherosclerosis. In one of these studies (69), plasma and urine proteomes were evaluated in 67 elderly patients with and without coronary artery disease (CAD), who presented typical angina symptoms as well as the classic risk factors for atherosclerosis (hyperlipidaemia, diabetes, history of smoking, familial history of CAD and hypertension). With a combination of 17 urinary polypeptides it was possible to discriminate between CAD positive and negative patients with 81% sensitivity, 92% specificity and 84% accuracy. It is interesting to note that this was not possible using plasma proteins. Some of these urinary proteins were identified as collagen α1 fragments (types I and III), and were later isolated from atherosclerotic plaques in the aorta of patients with this disease. Type III collagen fragments were also detected in urine of atherosclerotic mice (ApoE -/-) (70) indicating ECM remodelling in this animal model. Type I and type III collagens are among the most predominant proteins of the arterial extra cellular matrix and are important for arterial integrity and plaque stability. Degradation of collagen by matrix metalloproteinase’s (MMPs) is one of the main mechanisms of plaque rupture (71), therefore, it is logical to think that these fragments may be present in urine of patients with atherosclerosis. However, collagen α1 fragments (types I and III) have also been detected in urine of patients with diabetes (18), chronic kidney disease (19) and prostate cancer (9). Therefore, they are not indicative of a specific disease, but rather of a pathological state in general.
A large study of 623 patients with and without coronary artery disease (72) reported a panel of 238 proteins which were specific to patients with CAD (area under the ROC curve 87%). The panel of biomarkers included fragments of α1-antitrypsin, collagen α1 fragments (types I and III), granin-like neuroendocrine peptide precursor, membrane-associated progesterone receptor component 1, sodium/potassium-transporting ATPase gamma chain and fibrinogen- α chain. Biomarker expression also changed after long term treatment with irbesartan, returning to more normal levels. Another study published a couple of years later (73) aimed to find proteins involved in atherogenesis and therefore reflect the early stages of atherosclerosis, using the gold standard animal model of atherosclerosis, namely the ApoE (-/-) mice. The urinary proteomic profiles of ApoE (-/-) mice on control and high-fat diets were compared to wild-type mice on high-fat diet using MS. The results revealed 16 urinary polypeptides specific to ApoE (-/-) mice on the high-fat diet. These included fragments of α1-antitrypsin, epidermal growth factor (EGF), kidney androgen-regulated protein, and collagen. In a blind study the researchers were able to identify the atherosclerotic mice, with 90% sensitivity and 100% specificity, by detecting these protein biomarkers in urine. It is noteworthy that α1-antitrypsin, EGF and collagen type I were found not only to be highly expressed in atherosclerotic plaques of these mice but were also present in high concentrations in the urine of human patients with this disease. Therefore, it may also be possible to identify human patients with atherosclerosis using this panel of biomarkers.
These studies reflect the growing trend towards the search for a panel of biomarkers, which can yield greater sensitivity and specificity results than a single protein. Promising urinary protein biomarkers have been reported for cardiovascular disease, although to date none of these have been implemented in clinical practice.
Osteoarthritis
Osteoarthritis (OA) is a chronic, slow progressing condition characterized by the breakdown of cartilage in the joints. A diagnosis of OA is currently made based on medical history and clinical examination, and confirmed by measuring the joint space width using X-ray. However, with these diagnostic methods, it is impossible to detect the early metabolic changes that occur prior to the visible structural changes in the joint tissues; which raises the need for molecular biomarkers of OA.
Degradation of type II collagen by proteolytic enzymes secreted by chondrocytes and synoviocytes is a central feature of OA (74). Evidence suggests (75) that this degradation of the extra cellular matrix is mainly mediated by MMPs. Using LC-MS/MS, collagen type II peptides were isolated from urine of patients with the disease (76); fragments which were found to be mainly produced by MMP-13. This was later clinically validated (77) and the assay was shown to differentiate between patients with symptomatic OA and healthy individuals; therefore, type II collagen peptides are candidate biomarkers for MMP-13 activity in cartilage and the consequent pathogenesis of OA.
Another study (78) that compared 10 urine samples from women with severe OA and 5 healthy controls, using 2D-DIGE and mass spectrometry, revealed 13 proteins of interest that were significantly over expressed or under expressed in urine of OA patients compared to healthy controls. These proteins included β-actin, α1-microglobulin, apoptosis-inducing factor-2, Zn-α2-glycoprotein precursor, serpin β1 and β3, mannan binding lectin serine protease-2 precursor, kinninogen-1 precursor and α1-antitrypsin; however, this study focused on two specific sequences of fibulin-3 (Fib3-1 and Fib3-2) which were identified from the 2 most increased spots in the urinary proteome of the OA patients. Fibulin-3 is an extra cellular matrix protein found to be involved in the pathogenesis of OA via its association with Tissue Inhibitor of Metalloproteinase’s 3 (TIMP3) (79,80).
A global initiative has been put in place by the Osteoarthritis Research Society International (OARSI) (81) to assess 12 biochemical markers of osteoarthritis in serum and urine, examine how well they correlate with clinical outcome, and establish their predictive validity using a large data set. These biochemical markers include hyaluronan, COMP, collagen I and II epitopes, aggrecan and MMP-3. This initiative should accelerate biomarker discovery and development for OA, allowing a preventative approach for managing the disease and a better use of the existing therapies.
Concluding remarks
Current diagnostic biomarkers are suboptimal and of poor utility for low grade disease and surveillance. To date many potential urinary protein biomarkers have been explored for diseases such as prostate cancer, breast cancer, diabetes, osteoarthritis and kidney disease. Some of these biomarkers are detected early on in the asymptomatic stage of the disease and are useful for risk stratification or for early diagnosis, such as MMP-9 and ADAM-12 for breast cancer and Annexin A3 for prostate cancer. Other biomarkers, such as ephrin type-B receptor 4 and vitamin K-dependent protein Z in type 1 diabetes, are used in the later stages of the disease, but are not useful for the initial diagnosis. One of the biggest challenges is that biomarkers are often shared by several pathologies, so are not specific to one disease. It is not sufficient to compare the urinary proteomes of patients with a certain disease and healthy individuals. The key to identifying promising biomarkers is to compare multiple diseases (well-designed controls), to ensure high specificity. The trend is also shifting towards using a panel of biomarkers for a disease, rather than just one biomarker which may not be as specific. A panel of biomarkers will tolerate normal variability that is observed among individuals, and that is due to the heterogeneous nature of the disease, without jeopardizing the diagnostic precision. Some of the proteins present in these biomarker panels may not contribute significantly to the overall specificity and sensitivity. Therefore, it is also necessary to optimize these panels, so that enough biomarkers are included to ensure diagnostic precision, while keeping costs down avoiding testing for unnecessary biomarkers. Although there have been many advances in urinary proteomics, these have not resulted in similar advancements in clinical practice due to the difficulties mentioned above, which imply high costs and large data sets. In order to translate these potential biomarkers to clinical practice, vigorous validation is needed, with input from industry or large collaborative studies.
Acknowledgements
We acknowledge the funding received from the Instituto Tecnológico Metropolitano for project P10240.


